Down syndrome changes how an individual’s body and brain develop, which can cause mental and physical challenges later in life. However, outcomes vary greatly across individuals and their cognition, behavior, and social interactions. The molecular underpinnings of such variability remain unknown and could make a great difference for therapeutic interventions that can ameliorate outcomes for many individuals with Down syndrome. Understanding the molecular basis of such differences requires systematic mapping of the molecular pathways, biological processes, regulators, individual cells, and brain regions in the context of multiple individuals with Down syndrome with diverse outcomes.
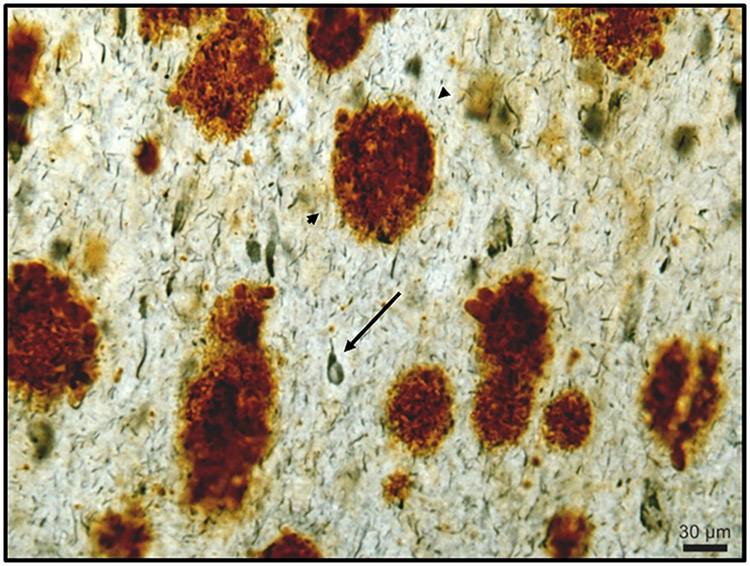
Due to an extra copy of the Alzheimer’s-related gene APP on chromosome 21, cognitive decline and amyloid plaque formation characteristic of Alzheimer’s disease are also greatly increased in prevalence across individuals with Down syndrome, providing an important and well-suited setting for studying phenotypic, molecular, cellular, and biological variability.
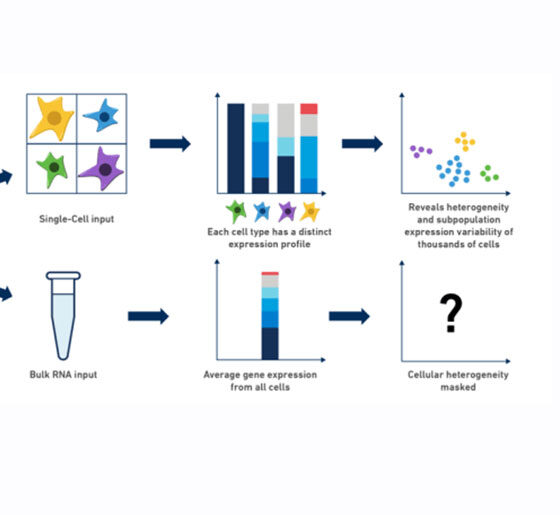
To address the heterogeneity of biological processes, cell types, brain regions, and individual cells across individuals with Down syndrome, Alana Center member Manolis Kellis has profiled 135,597 cells from 24 donated human postmortem samples with Down syndrome, some with and some without Alzheimer’s Disease. The team looked across two brain areas at the types and health of cells found in those regions, as well as the profile of gene expression found in those cells.
To do this, the team generates an ‘single-cell atlas,’ and labels and clusters cells into cell types, and then looks for genes that are regulated differently between the cell types. The lab also uses sophisticated computational methods to examine how gene expression is regulated in different cell types and states, with the goal of understanding the regulatory networks that control these. Gene regulatory networks are usually highly structured and regulated by a limited set of active regulators, which make them attractive candidates to approach as treatment targets that may cover many altered cellular processes.
Exciting preliminary discoveries
The lab has made some preliminary discoveries. They found that the presence of Alzheimer’s disease is associated with significant decreases in astrocytes and endothelial cells in a frontal cortical brain region. They observed down-regulation of DYRK1A, a gene previously linked to intelligence found on chromosome 21, in a subset of excitatory neurons in the amygdala.
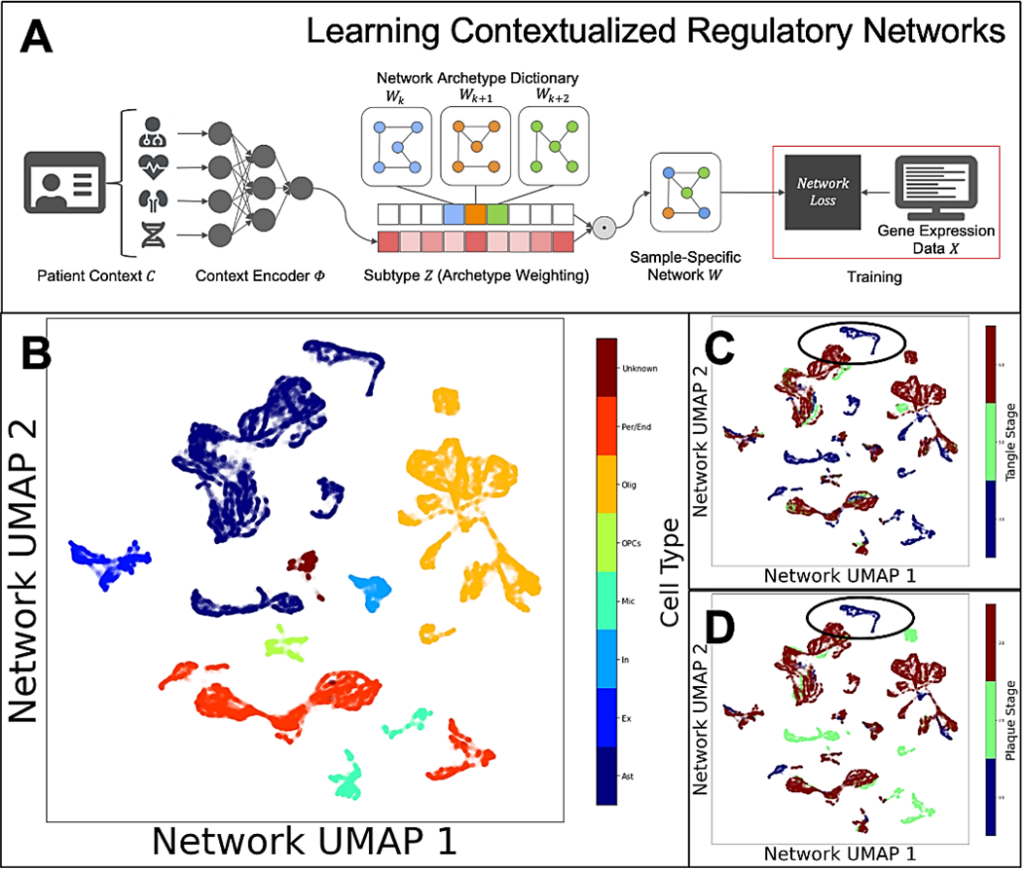
The team is also studying altered gene regulatory networks in astrocytes. Alana fellow Ben Lengerich developed computational methods to address inter-individual and inter-cell diversity by estimating gene regulatory networks influenced by patient-specific, tissue-specific, disease-specific, and cell-specific contexts and found that astrocyte networks cluster according to the onset of tangle and plaque formation.
Lastly, Alana fellow Bo Zhao and former fellow Michael Gutbrod developed high-throughput methods to alter gene regulators in iPSC-derived neurons and microfluidics-based spatial transcriptomics.