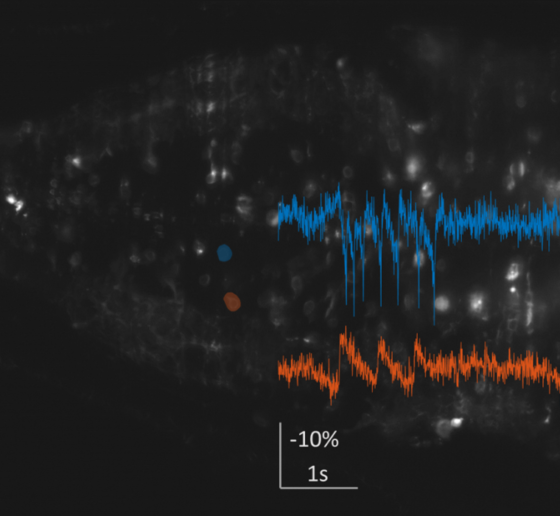
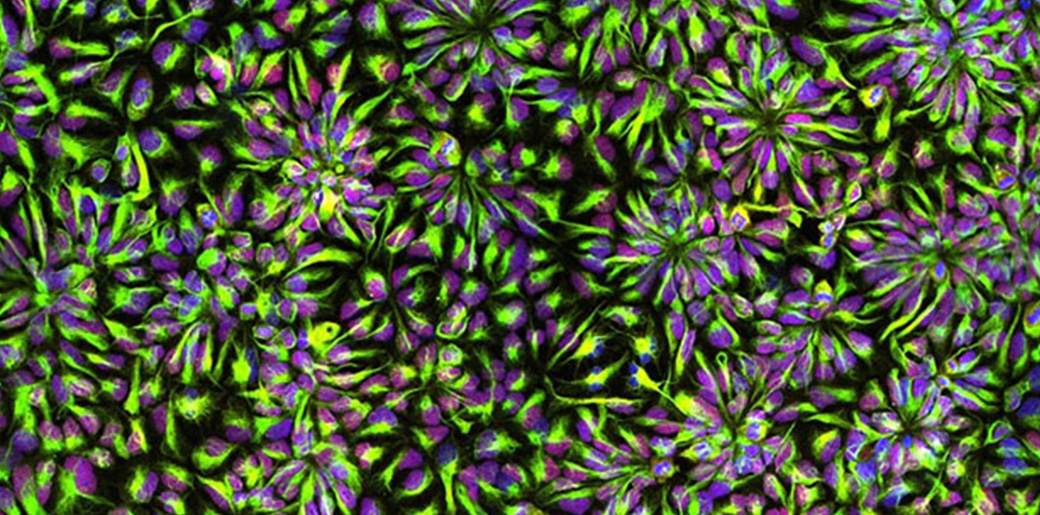
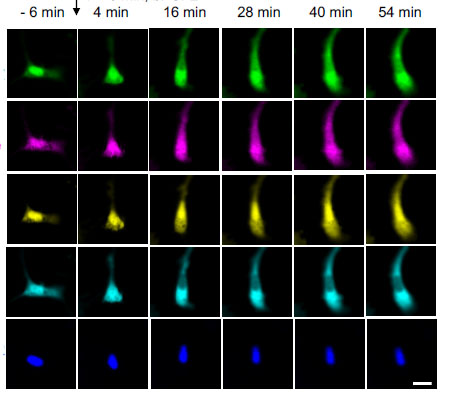
Imaging neurons as they fire
In a first line of work, the Boyden Lab has been developing imaging tools to characterize brain activity in Down syndrome model mice at the high speeds necessary to understand the mechanisms underlying the Tsai lab’s 40 Hz sensory stimulation protocol, GENUS. To do so, the lab has been identifying and developing new voltage indicators and creating new microscope architectures. The lab identified an excellent voltage indicator for in-animal neuron voltage imaging. To make high-speed, 3D imaging possible in the brains of freely moving mice, they are designing a miniaturized light-field endoscope that will allow simultaneous recording of neural activity in a 3-D volume at exceptionally high imaging rates. They have fabricated customized components, are working to finalize its construction, and are planning to apply it to studying Down syndrome mouse models in collaboration with the Tsai lab.
As part of this work, the Boyden lab recently identified sets of fluorescent probes that could be utilized simultaneously to do massively parallel imaging of many signals at once in a living cell, which is key to understanding how cells compute.
Expanding brains to see more clearly
Down syndrome involves changes in the organization of neural connections called synapses, which are too small scale to be imaged. In a second line of work, the Boyden Lab is refining its technology of expansion microscopy (ExM), which physically enlarges objects so they can be imaged with nanoscale precision on ordinary lab equipment.
They are developing a more powerful chemical method that locks proteins in place while enabling them to be cleanly separated from each other. The team is also creating a freezing protocol that uses cold temperatures to lock molecules in place while the aforementioned process is fixing them. The result is better preservation of delicate nanostructures. As part of this work, the team was able to publish their procedure on expanding proteins away from each other to greater extents than previously possible (see article on MIT news).